Memory Decoded? Scientists Simulate How the Brain Stores Information
A New Computational model reveals how memory-related proteins form unique structures vital for cognition and learning.
Ever wondered how a tiny thought can lead to experiences, learnings, and emotions? Or how does our brain store memories? The human brain is one of the most mysterious and complicated organs, and for ages, Scientists have been working tirelessly to decode its secrets and mysteries.
Recently, a team of researchers shed light on this fundamental mystery in Japan. An innovative and novel study reveals that the ‘proteins’ could be key in forming our beautiful memories.
These Researchers used an advanced Computational Model and simulated how proteins could organize as well as interact at synapses, thereby offering extraordinary insight into how the brain recalls as well as stores our memories. Could this be the key to our understanding of how we remember?
In a visionary effort, a team of Researchers from the ICBS (International Center for Brain Science), Fujita Health University, has developed a Computational model that simulates how some particular ‘memory-associated proteins’ self-organize themselves into highly specialized structures at the time of synapses. Their innovative study provides insights deep into the Biochemical and Physical processes that eventually give rise to unique ‘droplet-inside-droplet’ formations. These ‘droplet-inside-droplet’ formations are structures that could be critical to how we retain as well as learn information. Furthermore, they published this incredible study in ‘Cell Reports’ on 7th April, 2025.
This study gave insights and revealed proteins that shape memory, providing a novel perspective on how memories can be formed. So, are you excited and ready to explore the hidden building blocks of your mind? Then this is the perfect article for you!
Memory Mystery at the Molecular Level
Memory formation is widely believed to occur through the processes of adaptive changes in the strength and organization of synapses, as well as synaptic plasticity. Synapses are generally the communication points between our neurons. PSDs (Postsynaptic Densities), specialized protein-dense regions on the receiving side of synapses, are essential in these processes.
While earlier research has identified that the proteins within PSDs could cluster together during the process. In a 2021 Research study, it was highlighted that these proteins can assemble as well as bind into ‘droplet-like condensates’, membrane-less compartments similar to our Biological organelles. Moreover, these droplets possess an inner structure resembling a ‘droplet-inside-droplet’ organization. This smart observation raised quite critical questions, such as What governs their protein structure? How do they influence our memory? And why do such formations arise?
These interesting questions were tackled by a novel study driver by Dr. Vikas Pandey of ICBS (International Center for Brain Science), in partnership with Dr. Hidetoshi Urakubo from ICBS, as well as Dr. Yasunori Hayashi and Dr. Tomohisa Hosokawa from Kyoto University Graduate School of Medicine. Their research utilized advanced and futuristic Computational simulations to investigate as well as recreate the assembly of proteins at the molecular level (synaptic level).
Simulating Synaptic Protein Behavior
Focusing on four prime Synaptic proteins, the Research team paid attention to ‘CaMKII (Ca²⁺/calmodulin-dependent protein kinase II), which is a highly functionally significant as well as abundant protein in PSDs.
Ca²⁺/calmodulin-dependent protein kinase II is known for its important role in Synaptic Plasticity as well as its involvement in calcium signaling. Although its interactive and structural properties, especially in the context of memory formation, are less understood from a research perspective.
Leveraging futuristic Computational Modeling, the Research team recreated CaMKII as well as other important proteins’ interactions under various Biochemical conditions. Their simulations could successfully mimic the formation of the ‘droplet-inside-droplet’ and layered condensates observed in earlier experimental Research.
Figure: CaMKII Shape
The Key Mechanism Of The Research
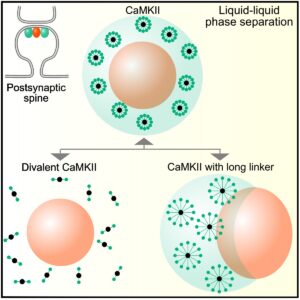
The model showcased a process – “LLPS” (Liquid-Liquid Phase Separation), which is a phenomenon where proteins form condensates without the need of any surrounding membrane.
Liquid-Liquid Phase Separation is emerging as a fundamental principle behind the organization of cellular components. This LLPS allows proteins to spontaneously aggregate into functional structures, just like oil droplets aggregate in water.
The Research revealed that LLPS leads to the formation of complex droplet structures as seen in memory-related synaptic proteins. These condensates aren’t unplanned but are influenced by the protein properties as well as the Biochemical forces.
One of the prime findings was that the characteristic shape of CaMKII, especially its short linker length as well as the number of binding sites (high valency), plays an essential role in these protein formations. These features generated conditions of reduced protein diffusion as well as low surface tension, aiding the proteins in maintaining a stable ‘droplet’ formation for extended periods. This condition was ideal for facilitating sustained synaptic signaling.
Dr. Vikas Pandey explained that “Our results revealed new structure–function relationships for CaMKII as a synaptic memory unit. This is the first systematic and mechanistic study investigating the divergent structure of protein-regulated multiphase condensates.”
He highlighted that the structural uniqueness of CaMKII makes it the perfect candidate for the formation of organized as well as long-lasting condensates at the synapses. These stable droplet structures could be instrumental in supporting the prolonged activation of Molecular pathways, which are essential for our memory formation.
“Droplet-Within-Droplet” Concept
The prime highlight of this study is its ability to demonstrate how and why the “droplet-inside-droplet” structures form, making it the differentiator. According to the Computational Model, these “droplet-inside-droplet” structures emerge due to competitive binding between the proteins. This binding is a situation where multiple protein types with varying binding affinities as well as shapes, interact simultaneously. CaMKII’s structure arranges as well as anchors other proteins into nested layers, acting as a “Molecular hub”.
The layered protein arrangement could eventually enhance the specificity as well as efficiency of memory-related Biochemical reactions.
Effects Beyond Neuroscience
While the immediate importance of this Research lies in understanding how memory functions at the Molecular level, its broader effects touch on a range of Psychiatric as well as Neurological conditions. Neurological Disorders like Autism Spectrum Disorders, schizophrenia, Down Syndrome, as well as Rett Syndrome, have all been linked to abnormalities in protein clustering as well as synapse formation.
Dr. Vikas Pandey further explained that the “Overall, the computational model developed in this study could serve as an important platform for investigating these conditions, potentially leading to new diagnostic tools and therapeutic approaches”.
Understanding how such protein condensates form as well as maintain their structure could unravel novel targets for early diagnosis, as well as Drug Development. Since LLPS-driven structures don’t depend on membranes, they could be more responsive as well as dynamic to environmental changes, ideal characteristics for therapeutic intervention.
Toward the Future of Brain Science
The development of this futuristic model represents a critical step forward in Neuroscience. By linking protein interaction and structure to synaptic functionality, the research bridges the gap between brain behavior as well as Molecular Biology.
Furthermore, the research utilized Computational simulations to complement experimental data and highlighted the increasing importance of Computational Neuroscience. As models become increasingly accurate, they can drastically reduce the need for trial-and-error in the research laboratories, expanding our understanding of the brain as well as speeding up novel discoveries.
This research unveils a fascinating mechanism underlying how memories might form at the Molecular level, but also opens the door to practical applications in Diagnostics as well as Medicine. As researchers continue to build on this work, we may soon be able to visualize how memories are stored, recalled, as well as encoded.
By doing so, we will step closer to decoding the most enigmatic organ in the human body: the magnificent brain!
Curious to unlock more secrets behind how your brain builds and stores memories? Explore the full Research Paper, where Life Science meets the subconscious, and the proteins tell numerous stories waiting to be deciphered!
Pandey, V., Hosokawa, T., Hayashi, Y., & Urakubo, H. (2025). Multiphasic protein condensation governed by shape and valency. Cell Reports. DOI: 10.1016/j.celrep.2025.115504