Next Generation Sequencing For Beginners
A Beginner’s Journey into NGS
The novel method of next-generation sequencing (NGS) has allowed for the rapid and efficient sequencing of DNA and RNA, radically altering the field of genomic research. An insightful summary of NGS is given by Dr. Nilofer, a bioinformatics expert at Biotecnika,who highlights the value of the method for finding unique variants, mutations and SNPs using reference genome sequences. Researchers can analyze millions of DNA fragments at once using NGS’s high-throughput capabilities, which set it apart from other sequencing approaches. This method is vital to modern genetic research since it has multiple applications in a wide range of disciplines, such as environmental science, agriculture and medicine. The development of NGS greatly improved the capacity to assess complex genomic data, leading to greater knowledge of genetic pathways and enabling the use of adapted treatment.
What is Sequencing?
The technique of determining the nucleotide order in nucleic acids (DNA/RNA) is known as sequencing. Genomic sequencing, which analyses genes and genomes, provides the basis for defining NGS. This method targets specific protein targets, analyzes changes in amino acid structure and finds possible drug targets using biological sequences in the form of DNA. The precise nucleotide order can be determined using a variety of methods in order to find changes or mutations.
DNA Sequencing
Based on ATGC combination, DNA sequencing determines the order of DNA nucleotides. The general makeup of an organism can be understood by using reference genomes. Sequencing was done using traditional methods like the one developed by Sanger using fluorescent dyes and the Maxam-Gilbert method using radioactive markers. Maxam-Gilbert method was considered obsolete as soon as Sanger’s method of DNA sequencing was developed. Sanger’s method caused a revolution in the molecular biology field and because of that Fredrick Sanger was even provided the Nobel Prize in 1980. But even Sanger’s method was time consuming and expensive, which called for the advent of more efficient approaches, like NGS.
Next Generation Sequencing (NGS)
Incorporating techniques like nanopore sequencing and anchored surface methods, NGS allows the sequencing of millions of amplified molecules each run. Large-scale sequencing is made feasible by these methods, which get beyond the drawbacks of traditional techniques. Given its high-throughput capabilities, NGS has altered sequencing by utilizing micro- and nanotechnology to handle smaller samples, need fewer reagents and produce faster results. Thousands of sequences can be widely in tandem sequenced at once using this technology, resulting in gigabyte-scale data quantities. Thus, NGS has the ability to produce enormous amounts of data while decreasing sample sizes, expenses and turnaround times.
Comparison of NGS with Traditional Sequencing
| Feature | Next Generation Sequencing (NGS) | Traditional Sanger Sequencing |
| Processing Capacity | High capacity, can sequence millions of reads simultaneously | Low capacity, reads one sequence at a time |
| Speed | Provides faster results due to parallel sequencing | Slower due to sequential reading |
| Scalability | Highly scalable for large-scale genomic studies | Less scalable, suited for smaller projects |
| Data Output | Generates large volumes of data | Produces smaller data sets |
| Cost | More cost-effective for high-throughput sequencing | More expensive for large-scale projects |
| Applications | Suitable for whole-genome sequencing, transcriptomics, and metagenomics | Ideal for small-scale projects and validation of NGS results |
| Accuracy | High accuracy, but validation with Sanger sequencing often required | High accuracy, used for confirmation of NGS data |
| Read Length | Typically shorter read lengths (50-300 bp) | Longer read lengths (up to 1000 bp) |
| Technology | Utilizes massive parallel sequencing platforms | Based on chain termination method |
| Flexibility | Flexible for various complex genomic studies | Limited flexibility, primarily used for specific tasks |
Opportunity to learn NGS from Basics to Advanced
Registrations Open For Next Generation Sequencing (NGS) Virtual Internship Program
Accelerate Your Research Career By Learning NGS in the comfort of your home.
Start Date: 12th August
Duration: 15 Days
Mode: Online
Next Generation Sequencing For Beginners
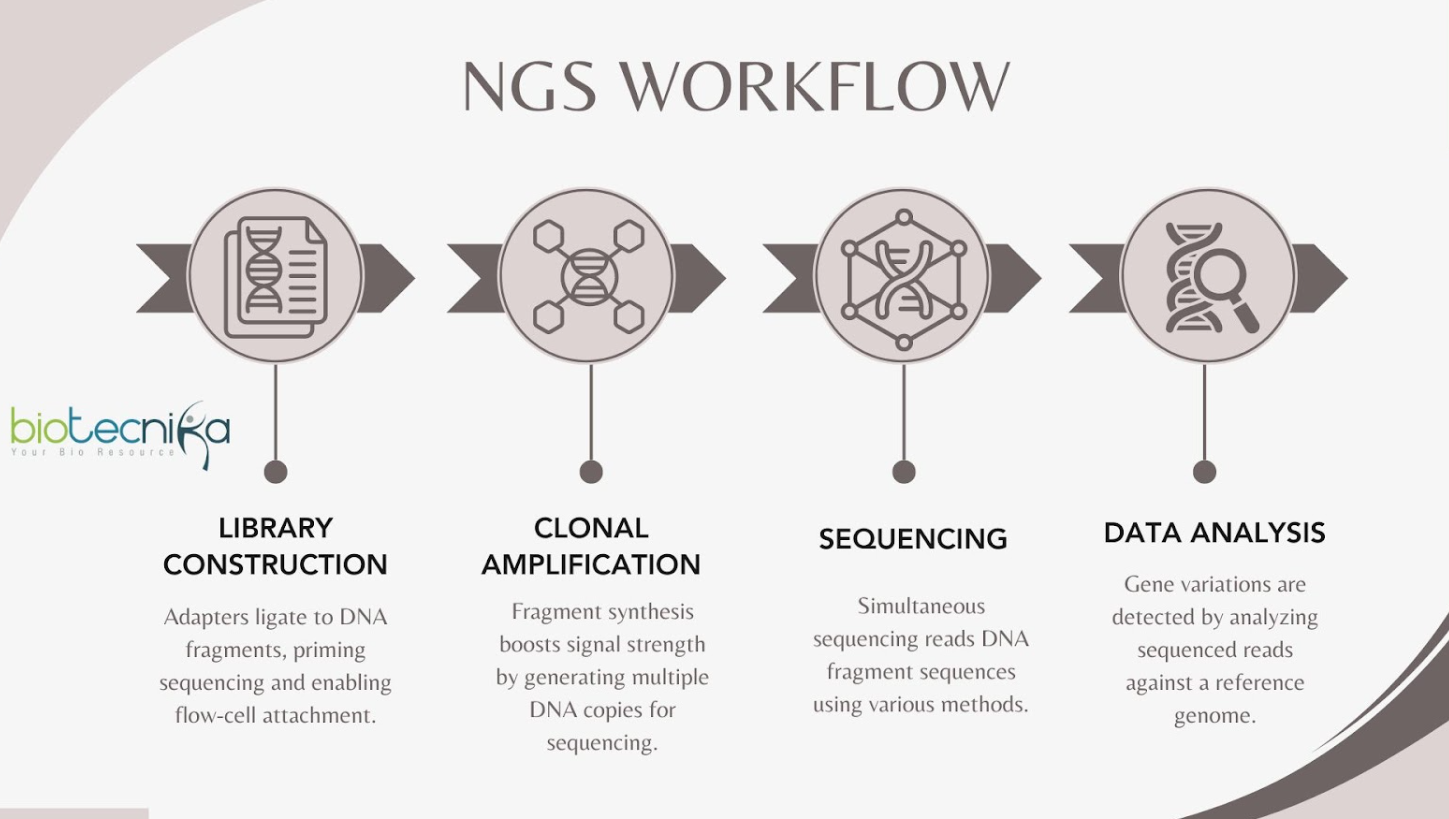
NGS Workflow
- Library Construction: The adapters are ligated to DNA fragments. These adapters, not only help in priming the sequencing process but are often marked with unique molecular barcodes for sample identification. These adaptors are also required for the DNA fragment to attach to the sequencing flow-cell.
- Clonal Amplification:Synthesis of the fragments increases signal strength. Multiple copies of each DNA fragment are generated in this stage to ensure an adequate signal for sequencing.
- Sequencing: Simultaneous sequencing is done on DNA fragments. In this step, the nucleotide sequences of the DNA fragments are read utilizing sequencing by synthesis or different methods.
- Data Analysis: Variations in genes are found via analysis of sequenced reads. Using a reference genome, bioinformatics techniques detect variations like SNPs and insertions/deletions.
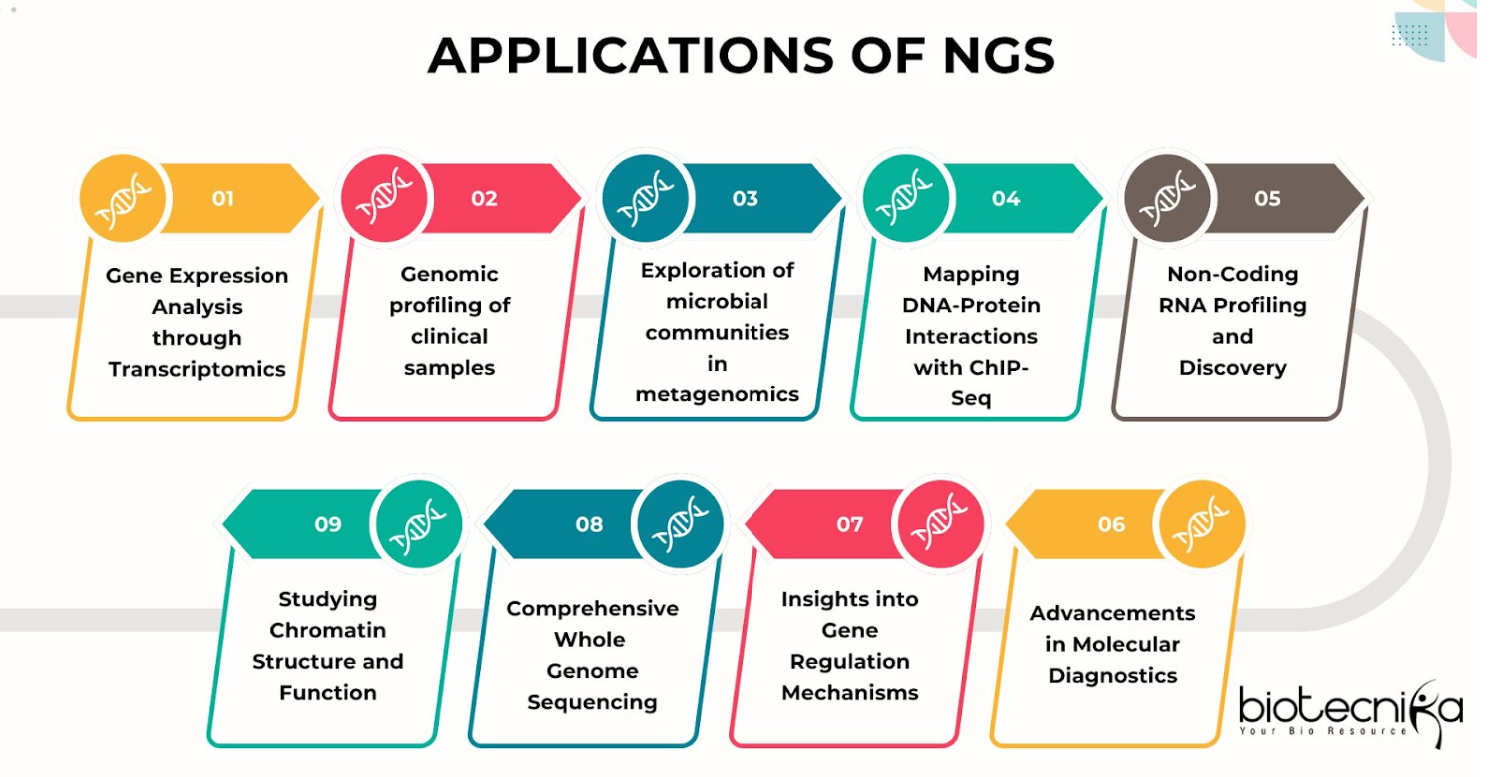
Applications of NGS
- Gene Expression Analysis through Transcriptomics: Studying gene activity through RNA sequencing. By examining the amounts of mRNA and discovering new RNA changes, RNA-Seq helps scientists learn new things about how genes are controlled.
- Genomic profiling of clinical samples: Examining differences in genes in medical samples. This aids in recognizing illnesses and finding their causes.
- Exploration of microbial communities in metagenomics: Studying microbial populations in different environments. Next-generation sequencing (NGS) is used in metagenomics to explore the diversity and functions of microbes in materials such as soil, water and the human digestive system.
- Mapping DNA-Protein Interactions with ChIP-Seq: Using ChIP sequencing to study interactions. This method helps us understand how proteins that attach to DNA, such as transcription factors, work in controlling genes.
- Non-Coding RNA Profiling and Discovery: Discovering and understanding non-coding RNAs. Next-generation sequencing helps find RNA molecules that don’t make proteins but are important for controlling the genome.
- Advancements in Molecular Diagnostics: Using Next-Generation Sequencing (NGS) in diagnosing cancer and inherited diseases. NGS helps in thoroughly examining the genetic makeup of cancer cells to find changes that cause tumors and possible treatment options.
- Insights into Gene Regulation Mechanisms: Studying gene regulation mechanisms. NGS techniques like RNA-Seq and ChIP-Seq provide detailed insights into how genes are turned on and off in different conditions.
- Comprehensive Whole Genome Sequencing: Sequencing entire genomes for comprehensive analysis. This application is essential for identifying genetic variations that contribute to complex traits and diseases.
- Studying Chromatin Structure and Function: Investigating chromatin structure and function. NGS helps in understanding how DNA is packaged within the nucleus and how this affects gene expression and genome stability.
Due to NGS, genome annotation and sequencing is now possible in a timely manner, revolutionizing the fields of molecular biology and genomic studies. Due to incorporation of NGS in various research fields, scientists are now able to learn more regarding genetic routes and gain insights which were not possible before with traditional approaches. With the use of this technology, personalized medicine can progress and treatments can be tailored to a patients genetic description, improving the results of therapy and reducing side effects. Additionally, NGS is very helpful in environmental studies, where it helps monitor ecosystem health and biodiversity. NGS can be applied in agriculture as well, where it helps by improving yields and resistance to diseases. NGS technology is a cornerstone of genetic and genomic research, and its constant enhancements promise even more growth.



































i am interested…