CSIR NET RNA Synthesis Notes – UNIT 3
UNIT 3 From CSIR NET Life Science Syllabus can help you fetch a good score in the exam. It’s an important unit and one must not skip it at all as it deals with molecular biology. From UNIT 3 one can expect an average of 12% questions in Part B and 8% questions in Part C.
Reference Books For UNIT 3B-RNA Synthesis & Processing:
1. Molecular biology, weaver, 5th edition
2. Principles of biochemistry, Lehninger, 6th edition
3. Molecular cell biology, Lodish, 8th edition
4. Watson, Molecular Biology Of The Gene, 7th Edition
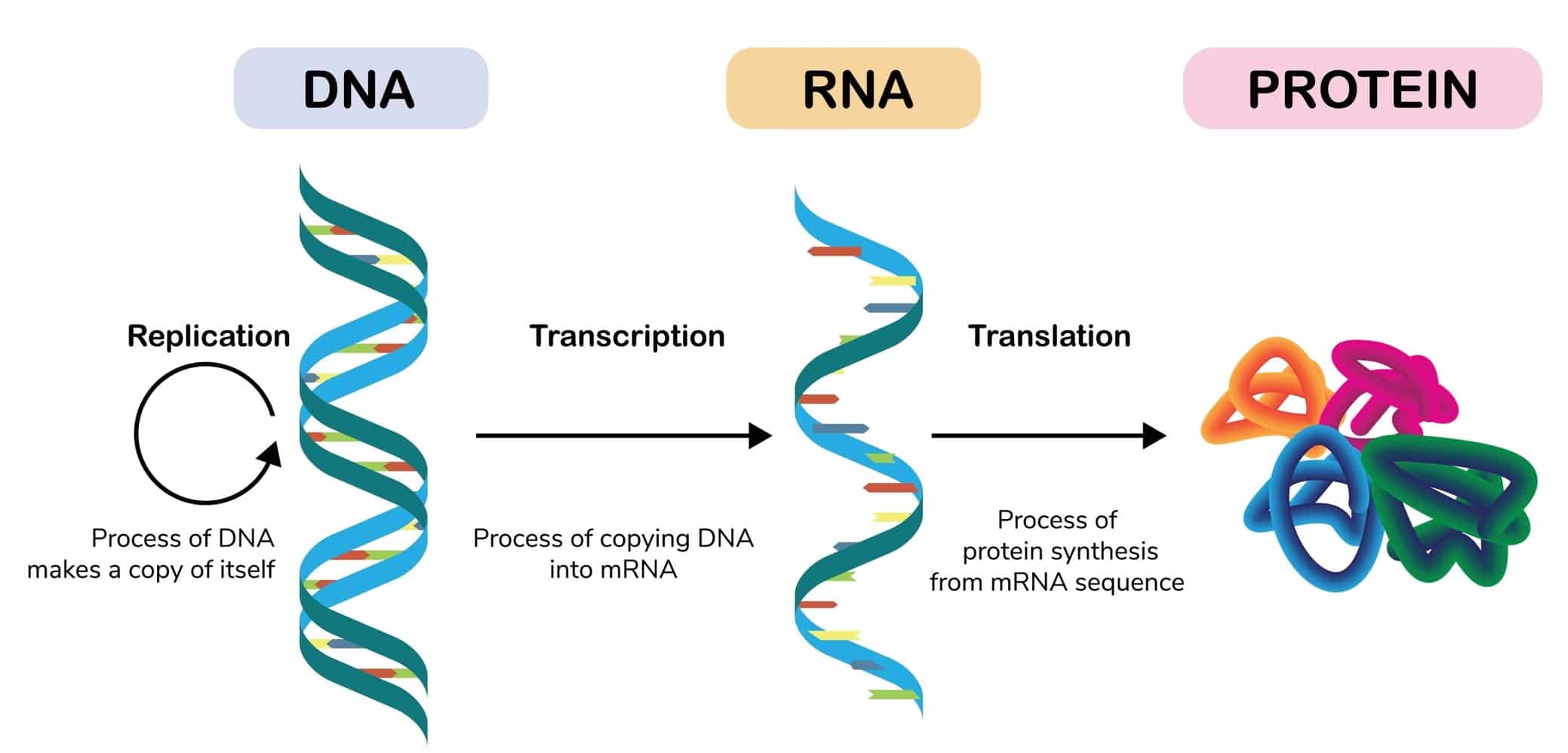
Central Dogma
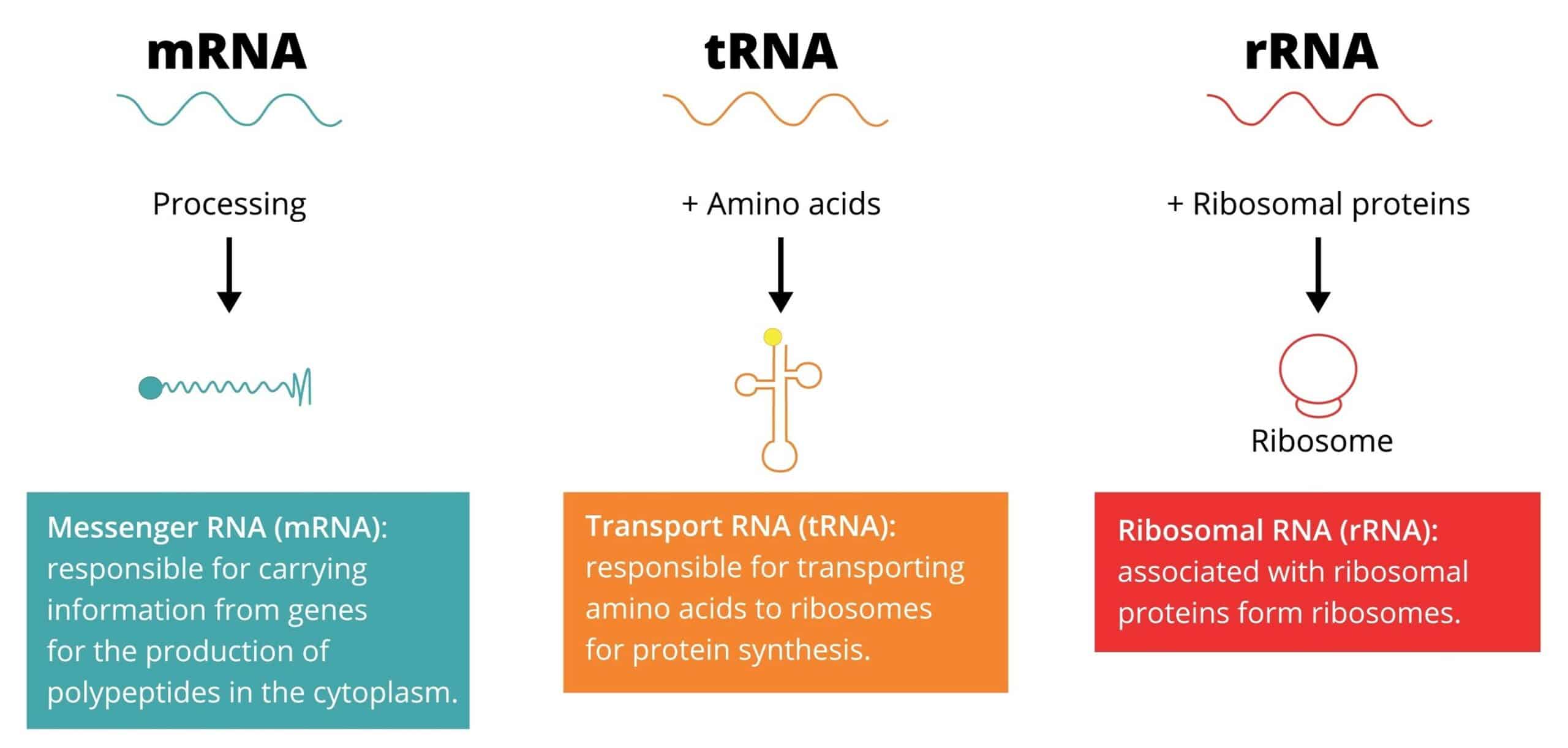
Post Transcriptional Processes
- The primary transcripts are not necessarily functional entities
- To acquire biological activity, many of them must be
specifically altered - Processes:
– Removal of a polynucleotide by the exo- and endonucleolytic
segments
– Appending nucleotide sequences to their 3’ and 5’ ends
– Modification of specific nucleosides
Structure of mRNAs
- A continuous sequence of nucleotides encodes a specific polypeptide.
- They are attached to ribosomes when they are translated.
- Most mRNAs contain a significant noncoding segment, that is, a portion that does not direct the assembly of amino acids.
- Eukaryotic mRNAs have special modifications at their 5’ and 3’ termini that are not found on either bacterial mRNAs or on tRNAs or rRNAs.
- The 3’ end of nearly all eukaryotic mRNAs has a string of 50 to 250 adenosine residues that form a poly(A) tail,
- Whereas the 5’ end has a methylated guanosine cap
Download Full PDF Notes For RNA Synthesis & Processing
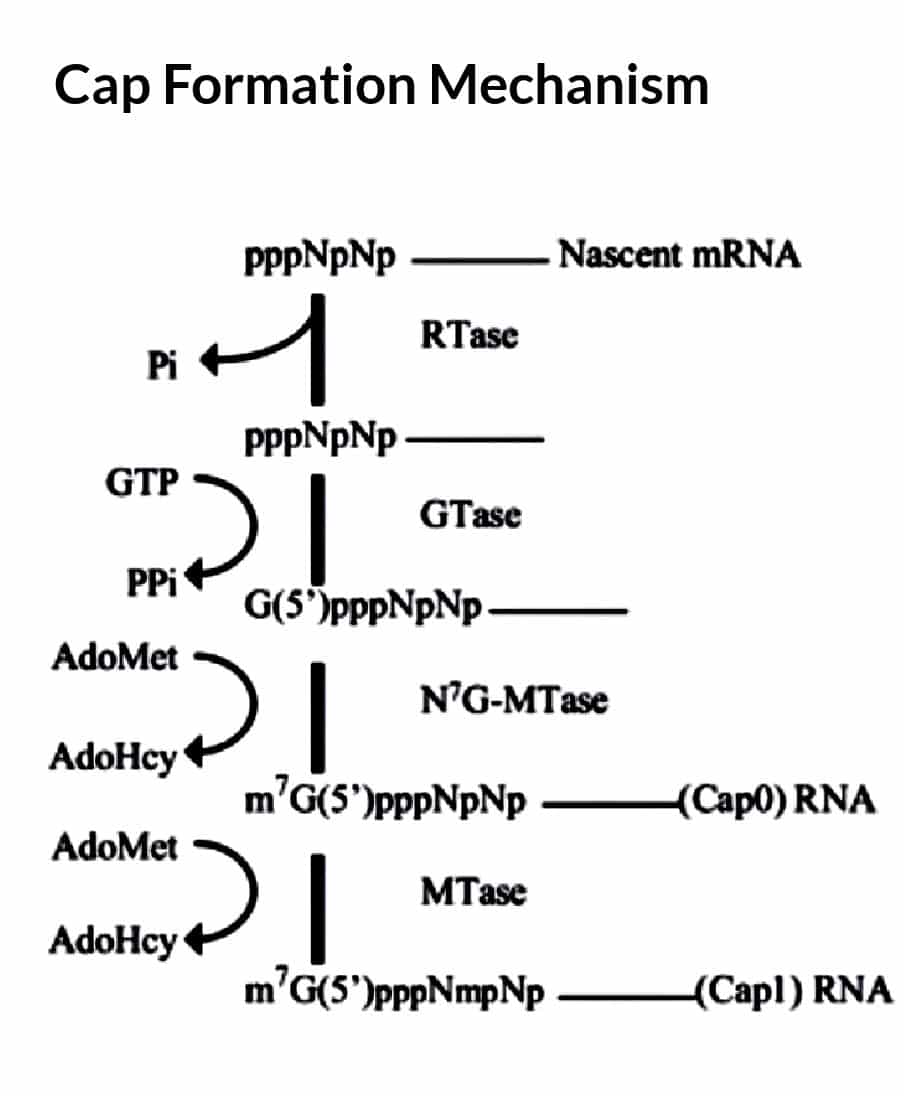
Cap
Eukaryotic mRNAs have a peculiar enzymatically appended cap structure consisting of a 7-
methylguanosine (m7G) residue joined to the transcript’s initial (5’) nucleoside via a 5’–5’ triphosphate bridge.
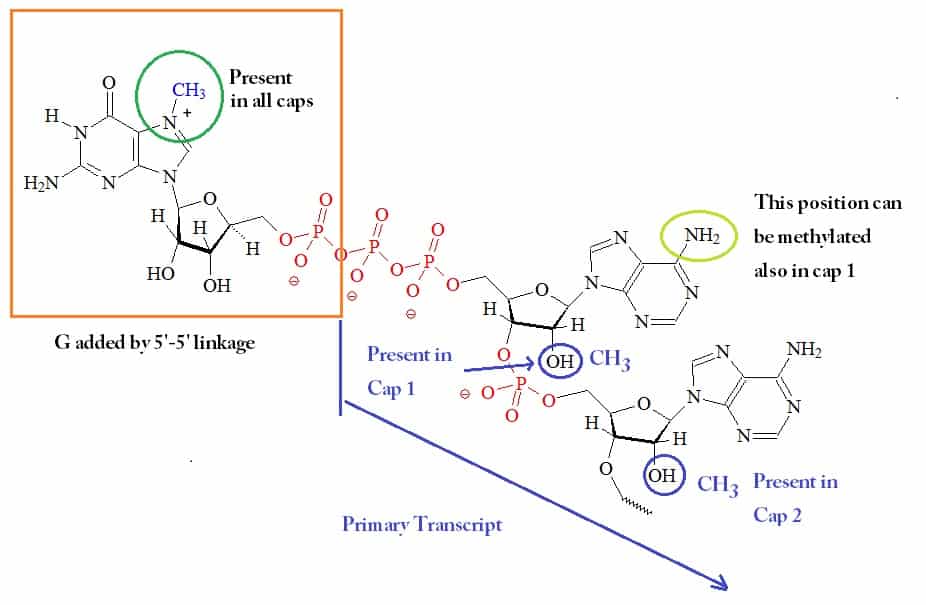
Functions
Caps appear to serve at least four functions:
- They protect mRNAs from degradation.
- They enhance the translatability of mRNAs.
- They enhance the transport of mRNAs from the nucleus into the cytoplasm.
- They enhance the efficiency of the splicing of mRNAs.
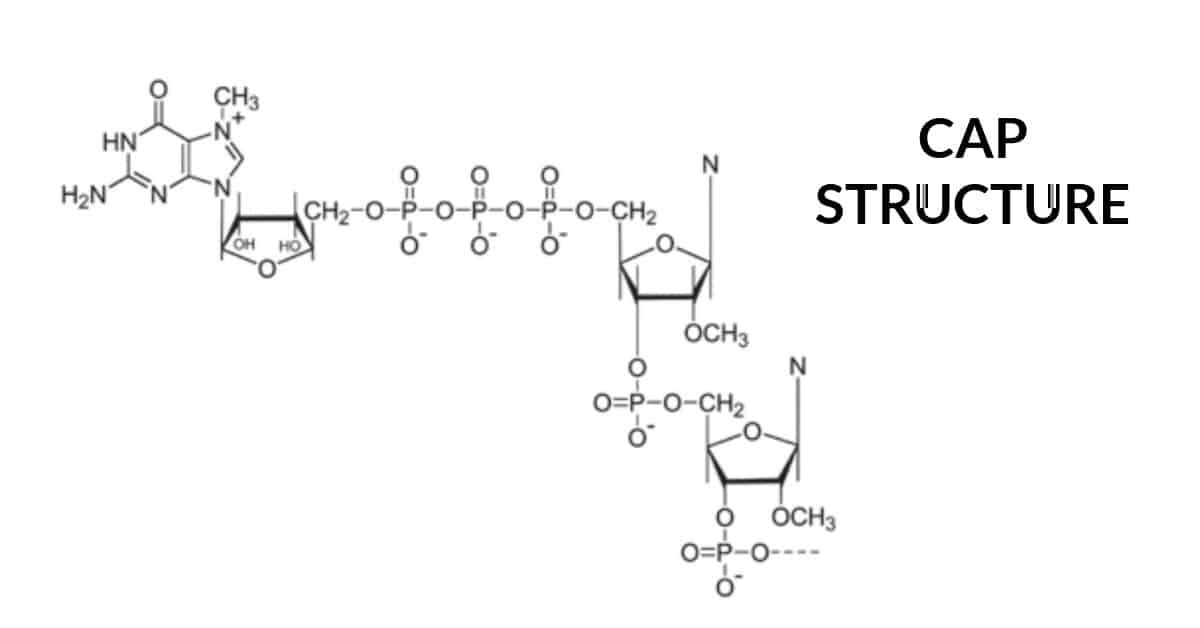
Process
- Phosphorylation by TFIIH at serine 5 residue (CTD of RNA pol II) recruits the capping
machinery.
- This m7 cap, which is added to the growing transcript before it is ̴30 nucleotides long, defines the eukaryotic translational start site
- Capping marks the completion of RNAP II’s switch from transcription initiation to elongation
- Enzymes involved – nucleotide phosphohydrolase (RNA triphosphatase), guanylyltransferase, methyltransferase
- First, an RNA triphosphatase removes the terminal phosphate from a pre-mRNA
- Next, a guanylyltransferase adds the capping GMP (from GTP).
- Next, methyltransferases methylate the N7 of the capping guanosine and the 2’-O-methyl group of the penultimate nucleotide.
Poly (A) Tail
- Mature eukaryotic mRNAs have well-defined 3’ ends; almost all of them in mammals
have 3’-poly(A) tails of ̴250 nucleotides
- Poly(A) enhances both the lifetime and translatability of mRNA. The relative importance of these two effects seems to vary from one system to another.
- Enzymatically appended to the primary transcripts in two reactions that are mediated by a 500- to 1000-kD complex that consists of at least six proteins.
- The mechanism of polyadenylation usually involves clipping an mRNA precursor, even before transcription has terminated, and then adding poly(A) to the newly exposed 3’-end
Polyadenylation Signals
Mammalian polyadenylation signal consists of an AAUAAA motif about 20 nt upstream of a polyadenylation site in a premRNA, followed 23 or 24 bp later by a GU-rich motif, followed immediately by a U-rich motif.
Polyadenylation Factors – Contd.
Download Full PDF – CSIR NET RNA Synthesis Notes
Detailed Notes on RNA Synthesis & Processing – Available including:
- RNA Synthesis & Processing Flow charts & Koncept Tables
- Multiple Lecture Videos on RNA Synthesis
- Q&A Sessions +
- Doubt Solving
Join CSIR NET + GATE Josh Batch December 2022 Starting on 6th June
Chat with us to Book your seat Today
Get a FREE Computational Biology Virtual Internship opportunity,
JCB Scholarships Available
Download CSIR NET Notes PDF
- UNIT 1(A) Notes – Structure of Atoms, Molecules & Chemical Bonds – https://btnk.org/unit1a
- UNIT 1(B) Composition, Structure & Function of Biomolecules – https://btnk.org/unit1b
- UNIT 1(G) Ramachandran Plot – https://btnk.org/unit1g
- UNIT 2 Organization of Genes & Chromosomes – https://btnk.org/unit2
- UNIT 2D Cell Division & Cell Cycle – https://btnk.org/unit2d
Keywords: CSIR NET RNA Synthesis Notes – UNIT 3, CSIR NET UNIT 3 Notes, CSIR NET Unit 3B RNA Synthesis & processing notes.




























 machinery.
machinery.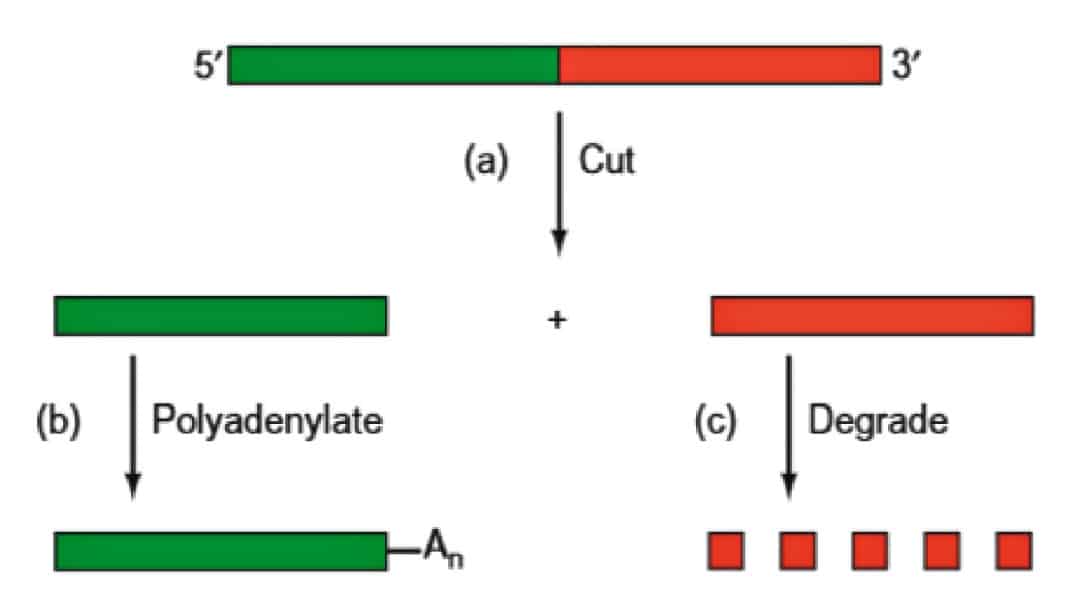 have 3’-poly(A) tails of ̴250 nucleotides
have 3’-poly(A) tails of ̴250 nucleotides









